CORE
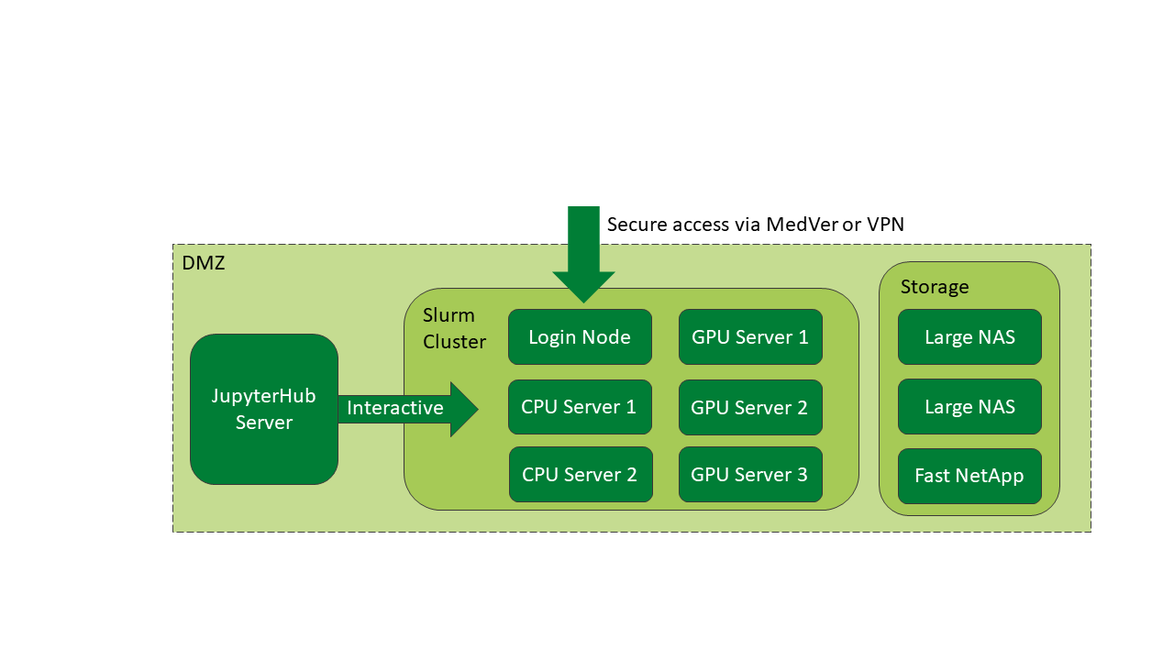
LMU Klinikum's High Performance Computing (HPC) infrastructure, known as CORE, is a collaborative HPC platform supporting scientific research within LMU Klinikum. Hosted by MIT, CORE is accessible via LMU Klinikum's MedVer network or remotely and securely through a VPN. Key components include a Slurm cluster with three GPU servers and two dedicated CPU servers, along with two spacious fileservers and a high-speed netapp. Additionally, CORE offers a jupyterhub instance for easily accessible interactive computing using jupyter notebooks, Rstudio and more.
CORE is a core facility of LMU Klinikum and a valuable resource for accelerating medical research, providing researchers from within LMU Klinikum with the computational tools needed to drive innovation and advance healthcare.
Slurm Cluster
To efficiently distribute the computing resources among the clinics and working groups of LMU Klinikum involved in compute-heavy research, CORE uses a scheduling software called Slurm. Slurm is a specialized, well-established HPC tool and ensures that computing workloads are scheduled on the hardware in a fair and efficient manner to maximize the utilization of the compute servers. Various Slurm partitions allow to efficiently distribute the resources to GPU- and CPU-only-jobs and to accommodate jobs of different run times from quick prototyping to week-long deep-learning training. Up to eight modern GPUs within one node enable data-distributed parallel training runs. The container engine singularity (apptainer) allows users to package and ship their own libraries and computing environments on CORE.
JupyterHub Server
Very often, the focus of scientific computing is not heavy compute workloads, but interactive development, coding, debugging, statistical analysis or visualization. For these purposes, we provide a jupyterhub instance, backed by the compute and storage resources of CORE.
On this instance, researchers can open JupyterLab and RStudio environments from their browsers and have web-based access to their data and results stored on CORE. They can also interactively develop their code or run evaluations on their results.

How to join
As a core facility of LMU Klinikum, CORE is accessible for all research groups of LMU Klinikum in a bottom-up approach. In addition, guest researchers and collaboration partners can access CORE, as long as they have an official affiliation with LMU Klinikum - e.g. based on a "Hospitationsvertrag". If you are interested in joining CORE, please reach out to the steering committee (michael.ingrisch@med.uni-muenchen.de). For details, we refer to the Nutzungsordnung.
Documentation
We have put considerable effort into providing user documentation on CORE. All docs are hosted on LRZ's gitlab instance and can be viewed when logged into GitLab at CORE Documentation.

All documentation lives in a GitLab repository - and this enables user participation in a very straightforward way. Whenever you find bugs, typos, or want to extend the documentation with a tutorial, an example or anything else, you can contribute your content with a merge request.
Workshops
On a regular basis introductory workshops are hosted by the team of CORE, to equip users with the skills and knowledge to run their code on the cluster. Target audience of the workshop are researchers comfortable running their programs on the commandline, previous experience of working on HPC environments are not needed. The workshop is open to everyone interested, access rights to CORE are necessary for participation and the registration by email required. Please contact core.support@med.uni-muenchen.de to register or to be placed on the waiting list for the next workshop.
Our workshops are scheduled on demand, please let us know if you are interested. Workshops are also announced on the CORE mailing list.
Past workshops took place:
- March 22
- March 23
- October 23
- April 24
- November 24
- April 25
- November 25
Once you have access to the documentation, you'll also be able to consult the most recent workshop slides:
https://core.pages.gitlab.lrz.de/core-slides/
You can register to the next workshop here: https://www.med.moodle.elearning.lmu.de/course/view.php?id=8221
Nutzungsordnung
Stand: 13.10.2025
CORE is a joint effort with numerous contributors throughout the entire LMU Klinikum:
- Klinik und Poliklinik für Radiologie
- Klinik und Poliklinik für Dermatologie und Allergologie
- Klinik und Poliklinik für Psychiatrie und Psychotherapie
- Medizinische Klinik und Poliklinik I
- Kinderklinik und Kinderpoliklinik im Dr. von Haunerschen Kinderspital
- Dekanat der Medizinischen Fakultät
- Abteilung für Medizintechnik und IT (MIT)
The governance of the CORE core facility is determined by the Nutzungsordnung and comprises a steering committee and a user board.
Technical Administration Team
The technical administration team meets in a regular jour fixe, optimizes and adjusts infrastructure, and ensures day-to-day administration and maintenance.
Team members:
- Gabriel Lindner, Stabsstelle Digitale Medizin
- Dr. Balthasar Schachtner, MIT
- Simon Leutner, MIT
- Prof. Dr. Michael Ingrisch, Group leader Clinical Data Science, Department of Radiology
Contact: core.support@med.uni-muenchen.de
List of publications that have benefited from CORE:
2025
Stüber, A. T. et al. Replication study of PD-L1 status prediction in NSCLC using PET/CT radiomics. European Journal of Radiology 183, 111825 (2025).
Grosu, S. et al. Effect of artificial intelligence-aided differentiation of adenomatous and non-adenomatous colorectal polyps at CT colonography on radiologists’ therapy management. Eur Radiol 1–9 (2025) doi:10.1007/s00330-025-11371-0.
Hoelz, H. et al. Persistent desmoglein-1 downregulation and periostin accumulation in histologic remission of eosinophilic esophagitis. J Allergy Clin Immunol 155, 505–519 (2025).
Böck, A. et al. An integrated molecular risk score early in life for subsequent childhood asthma risk. Clin Exp Allergy https://doi.org/10.1111/cea.14475 (2024).
Longo, C. et al. Are single-nucleotide polymorphisms previously linked to inhaled corticosteroid response associated with obese-asthma in children? Pediatr Allergy Immunol 35, e14279 (2024). https://doi.org/10.1111/pai.14279
2024
Vetter, C. S. et al. Exploring the Predictive Value of Structural Covariance Networks for the Diagnosis of Schizophrenia. Preprint at https://doi.org/10.31219/osf.io/twgjb (2024).
Avraham-Davidi, I. et al. Spatially defined multicellular functional units in colorectal cancer revealed from single cell and spatial transcriptomics. 2022.10.02.508492 Preprint at https://doi.org/10.1101/2022.10.02.508492 (2024).
Ishar, J., Tam, Y. M., Mages, S. & Klughammer, J. BoReMi: Bokeh-based jupyter-interface for registering spatio-molecular data to related microscopy images. PLOS Computational Biology 20, e1012504 (2024).
Mittermeier, A. et al. Automatische ICD-10-Codierung. Radiologie 64, 793–800 (2024).
Ingrisch, M. et al. Automated Lesion Segmentation in Whole-Body PET/CT - Multitracer Multicenter generalization. (2024) doi:10.5281/zenodo.10990932.
Koutsouleris, N. et al. Distinct multimodal biological and functional profiles of symptom-based subgroups in recent-onset psychosis. Preprint at https://doi.org/10.21203/rs.3.rs-3949072/v1 (2024).
2023
Weber T, Ingrisch M, Bischl B, Rügamer D. Constrained Probabilistic Mask Learning for Task-specific Undersampled MRI Reconstruction. IEEE/CVF Winter Conference on Applications of Computer Vision (WACV 2024). 2024.
Weber T, Ingrisch M, Bischl B, Rügamer D. Cascaded Latent Diffusion Models for High-Resolution Chest X-ray Synthesis. In: Advances in Knowledge Discovery and Data Mining: 27th Pacific-Asia Conference, PAKDD 2023. 2023.
Weber T, Ingrisch M, Bischl B, Rügamer D. Implicit Embeddings via GAN Inversion for High Resolution Chest Radiographs. In: Medical Applications with Disentanglements. MAD 2022. Lecture Notes in Computer Science, vol 13823. Springer, Cham. 2023 Feb 1. doi: 10.1007/978-3-031-25046-0_3

